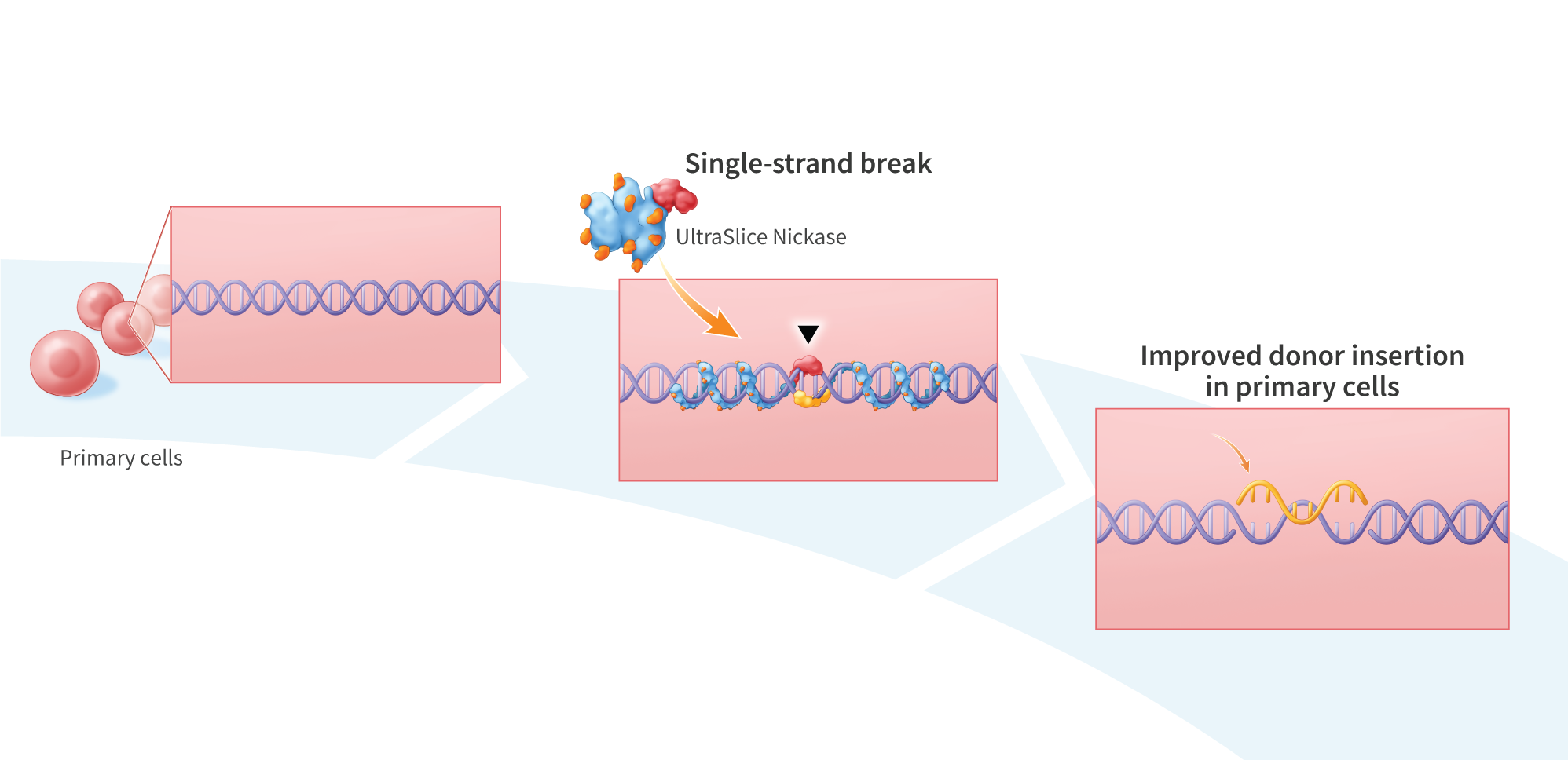
Gene Editing Proteins with Nickase Functionality Enable Scarless Targeted Gene Insertion in Primary Human Cells
Many gene editing strategies involve the use of nucleases to generate targeted double-strand breaks (DSBs) in genomic DNA, which is often associated with cytotoxicity and off-target effects that can prevent clinical translation. Such undesirable outcomes have led to the development of gene-editing nickases, which instead create targeted single-strand breaks (SSBs) that favor high- fidelity repair through the homology-directed repair (HDR) pathway rather than the more error-prone non-homologous end joining (NHE]) pathway. Here, we explore the use of UltraSlice gene editing proteins containing cleavage domain variants with nickase functionality for targeted insertion of donor sequences into a defined genomic locus. Using UltraSlice gene editing proteins targeting exon 73 in COL7A1 (mutations in which cause dystrophic epidermolysis bullosa), we tested combinations of 3 mutations previously reported to confer nickase functionality to the catalytic domain of Fokl, a Type IIS restriction endonuclease (D450A, D450N, and D467A). Notably, we found that D450A and D450N resulted in significantly reduced NHEJ relative to the COL7A1_e73 UltraSlice pair with a wild-type FokI cleavage domain, with D450N exhibiting the least amount of NHE]. We confirmed these results with Sanger Sequencing, comparing the D450N-treated PCR amplicon to a wild-type COL7A1 PCR amplicon and observed no significant alteration of the genomic target site. We then compared the ability of nickases to insert a 300 bp dsDNA donor sequence via electroporation into primary human fibroblasts. Insertion band intensities showed high insertion efficiencies for D450A and D450N (58.7% and 42.9%, respectively), though lower than standard UltraSlice (72.3%). Our data demonstrate that gene-editing nickases enable scarless insertion of donor sequences into defined genomic loci, and thus may have the potential to improve the safety ofin vivo gene insertion by reducing off-target effects.